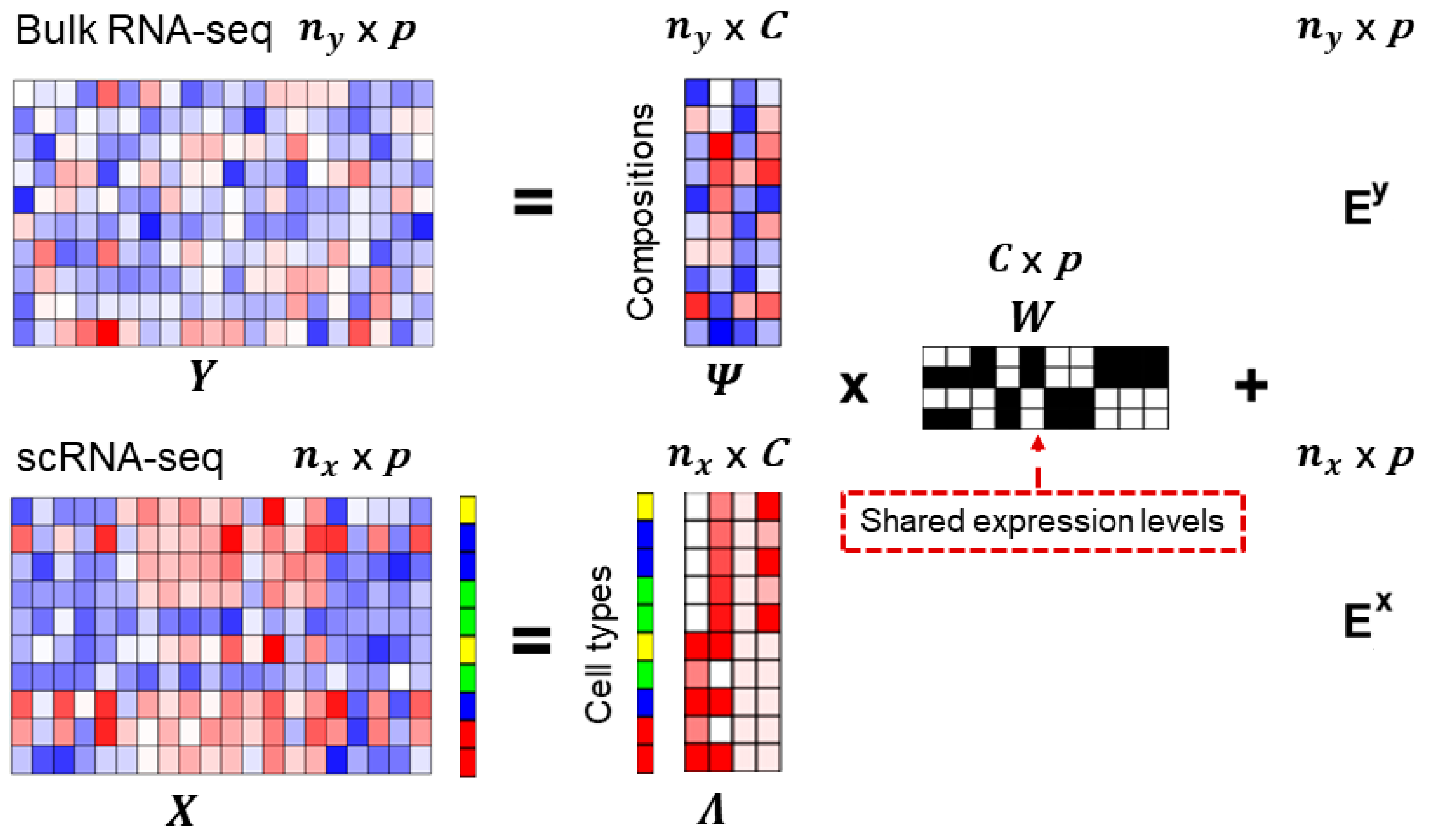
Cells | Free Full-Text | An Efficient and Flexible Method for Deconvoluting Bulk RNA-Seq Data with Single-Cell RNA-Seq Data

CDSeq – A novel complete deconvolution method for dissecting heterogeneous samples using gene expression data | RNA-Seq Blog

Breathing fresh air into respiratory research with single-cell RNA sequencing | European Respiratory Society

Illustration of popular methods used to analyse bulk and single-cell... | Download Scientific Diagram
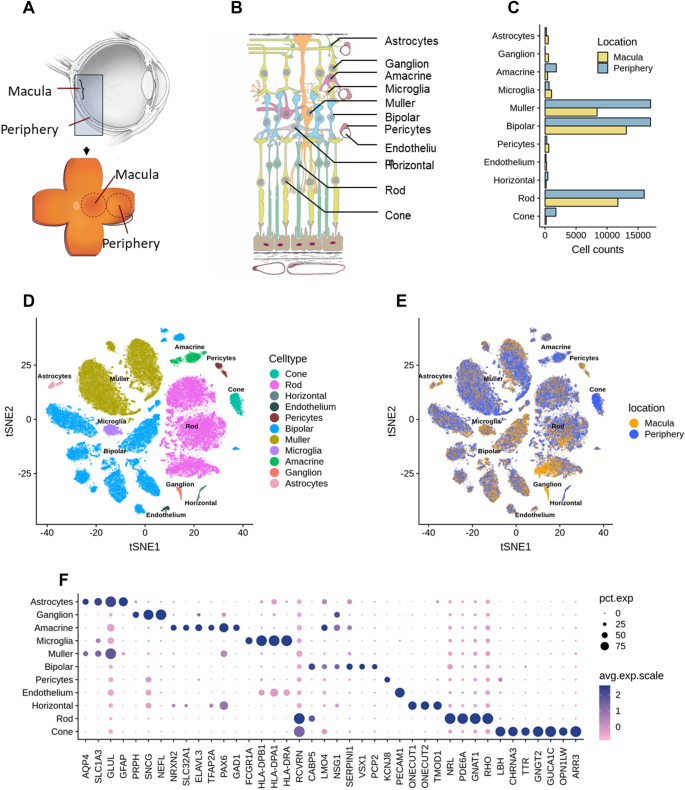
Implication of specific retinal cell-type involvement and gene expression changes in AMD progression using integrative analysis of single-cell and bulk RNA-seq profiling | Scientific Reports

Integrative single-cell and bulk RNA-seq analysis in human retina identified cell type-specific composition and gene expression changes for age-related macular degeneration | bioRxiv
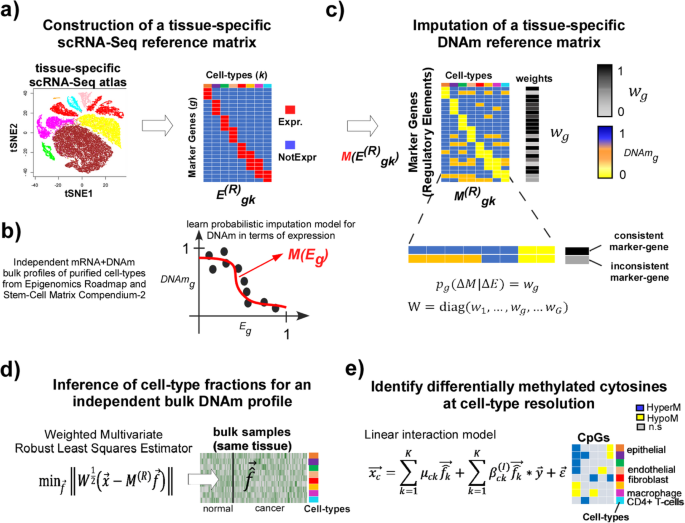
EPISCORE: cell type deconvolution of bulk tissue DNA methylomes from single- cell RNA-Seq data | Genome Biology | Full Text
Detecting cell-type-specific allelic expression imbalance by integrative analysis of bulk and single-cell RNA sequencing data | PLOS Genetics

Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants | Nature Communications

A protocol to extract cell-type-specific signatures from differentially expressed genes in bulk-tissue RNA-seq: STAR Protocols
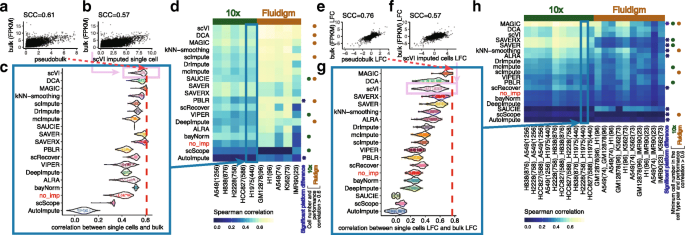
A systematic evaluation of single-cell RNA-sequencing imputation methods | Genome Biology | Full Text
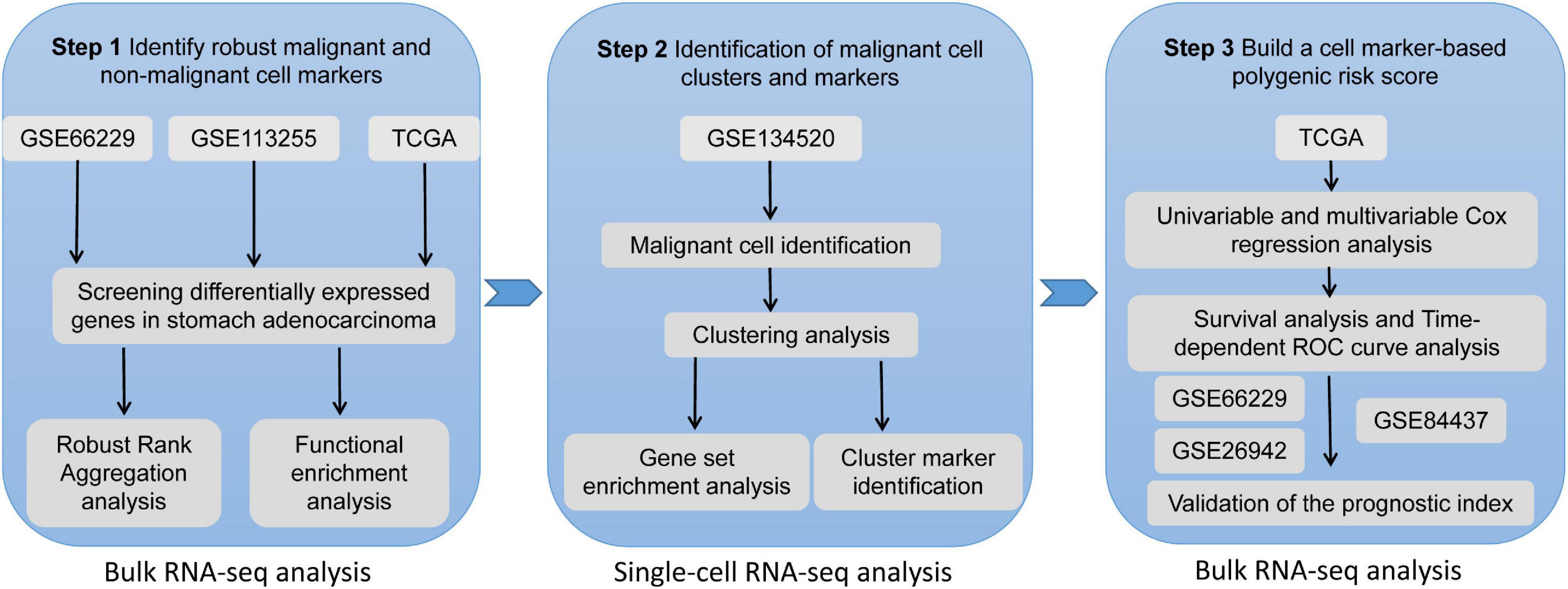
Frontiers | Identification and Validation of a Malignant Cell Subset Marker-Based Polygenic Risk Score in Stomach Adenocarcinoma Through Integrated Analysis of Bulk and Single-Cell RNA Sequencing Data
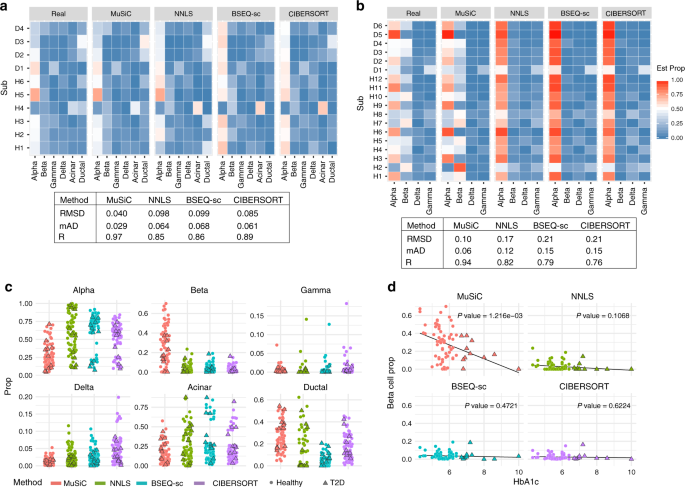
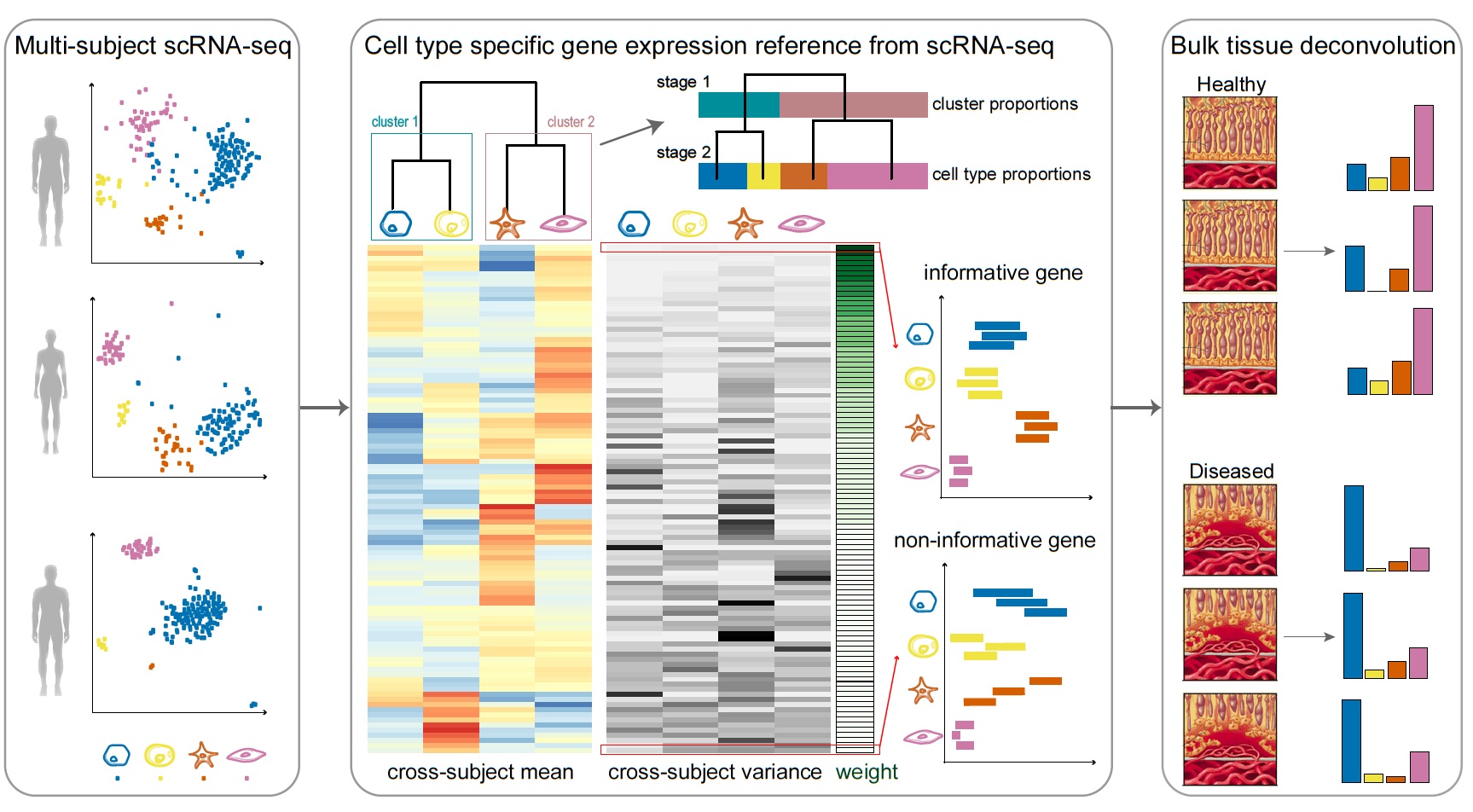

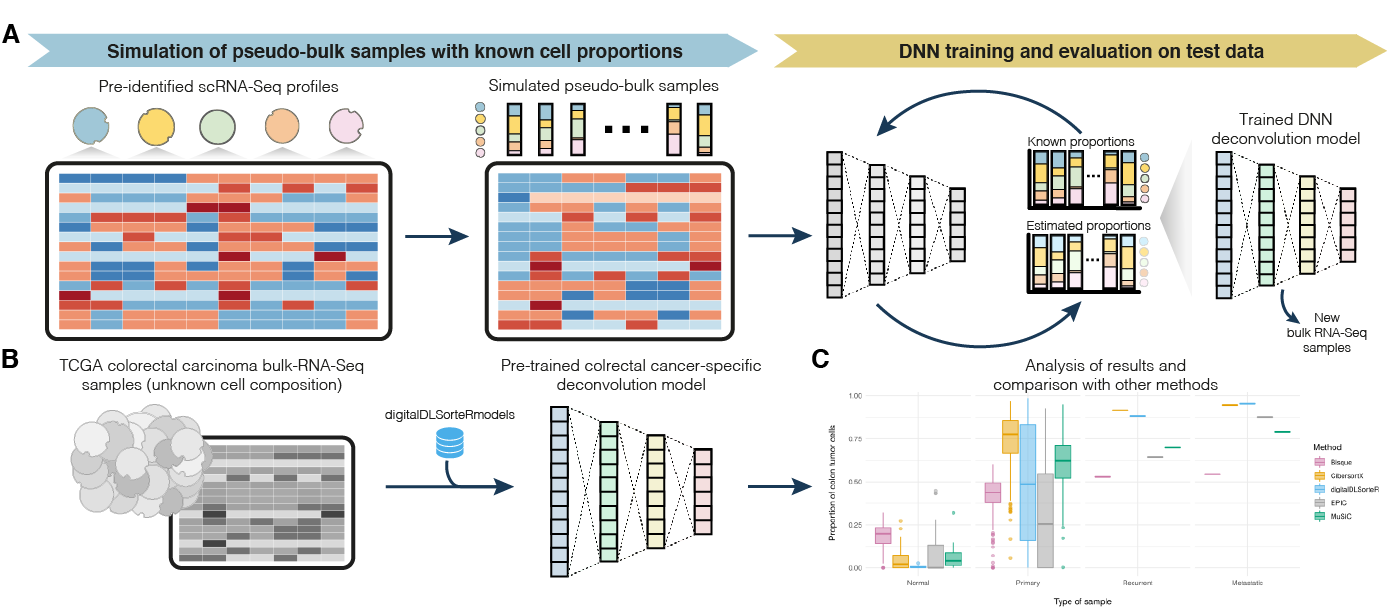




![BingleSeq: a user-friendly R package for bulk and single-cell RNA-Seq data analysis [PeerJ] BingleSeq: a user-friendly R package for bulk and single-cell RNA-Seq data analysis [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/10469/1/fig-1-2x.jpg)